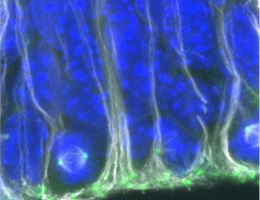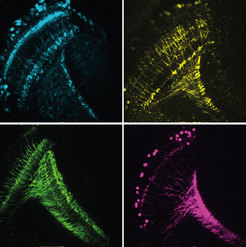
In order to react to changes in the environment in good time, the brain must analyze the signals it receives from the eyes rapidly and accurately. For example, the ability to recognise the direction in which an approaching car is moving is vital to the survival of modern humans in cities. Using the brain of the fruit fly Drosophila as a model, scientists from the Max Planck Institute of Neurobiology study how the brain extracts this essential motion information. They have now described in detail the cells that enable downstream neurons to recognize the direction of movement. Interestingly, the characteristics of these input cells exactly match to a motion detector model they recently proposed. In addition, the cells alter their characteristics according to the animals’ state: when the fly is active, the cells respond faster to light stimuli.
Humans perceive their environment mainly through their eyes. The ability to recognize movements and their direction is something that seems almost trivial and automatic to us. However, this information has to be processed in the brain as the light-sensitive sensory cells of the retina can only register changes in contrast. The direction of a movement can only be calculated through the comparison of neighbouring signals. Various models exist for these calculations. Alexander Borst and his team at the Max Planck Institute of Neurobiology study the extent to which these models can be applied to the brain’s neuronal circuitry. Their test subject is the fruit fly Drosophila, a master of motion perception.