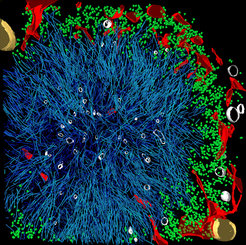
A common feature of neurodegenerative diseases such as Alzheimer’s, Parkinson’s, and Huntington’s is the accumulation of toxic protein deposits in the nerve cells of patients. Once these aggregates appear, they begin to proliferate like weeds. If and how these deposits damage nerve cells and lead to their demise remains largely unexplained. A detailed insight into the three-dimensional structure of the protein aggregates should help researchers to solve this puzzle. Now, using cryo-electron tomography, scientists at the Max Planck Institute of Biochemistry in Martinsried near Munich have succeeded in generating a high-resolution, three-dimensional model of the huntingtin aggregates responsible for Huntington’s disease. The results are published in the journal Cell.
Rampant weed growth – the nightmare of every hobby gardener. Trimming, cropping, cutting. Thorough garden maintenance is required. If this maintenance is neglected, weeds gain the upper hand and suppress the growth of crop and ornamental plants. The same applies to proteins in our bodies: molecular machines, large protein complexes that control vital cellular processes, assume the responsibility of a gardener. These molecular machines ensure that proteins reach their correct conformations and tend to and care for them for the duration of their lifespans.
A matter of the correct form
In order to carry out its function, a protein needs to adopt its correct three-dimensional structure. The building blocks of proteins, the amino acids, are assembled into long chains and folded into a complex form. If the resulting structure is faulty, the defective proteins are broken down in a strictly regulated process. If this does not occur properly, the misfolded proteins may aggregate forming clumps and deposits. Insoluble protein aggregates are toxic for cells. In the brain of patients suffering from neurodegenerative diseases such as Alzheimer’s, Parkinson’s, or Huntington’s, protein aggregates are often found.
If and how exactly these aggregates exert their toxic effects has not yet been explained. This is the question studied by the ToPAG (Toxic Protein AGgregation in neurodegeneration) consortium. A team of researchers in the departments of Wolfgang Baumeister, Ulrich Hartl and Rüdiger Klein has succeeded in decoding a 3D structure of the protein aggregates linked to Huntington’s disease within their intact cellular environment.
read more