Messenger RNAs (mRNAs) are the functional link between the genetic information in the cell nucleus and ribosomes, where proteins are synthesized. The structure of mRNAs can be differentiated into translated and untranslated regions. The translated regions serve as templates for the synthesis of proteins, while the untranslated regions have regulatory functions. The untranslated regions of mRNAs of all higher developed cells – from yeast to plants and humans – contain similar characteristic elements. One such element is the poly(A)-tail: a long chain of adenine molecules, one of the RNA building blocks. These tails are added to the end of the mRNA after their synthesis and fulfil many functions e.g. control stability, translation into proteins and localization of the mRNA. Nascent mRNAs have a long tail of up to several hundred adenines, which is then reduced to a species-specific length by enzymes called deadenylases.
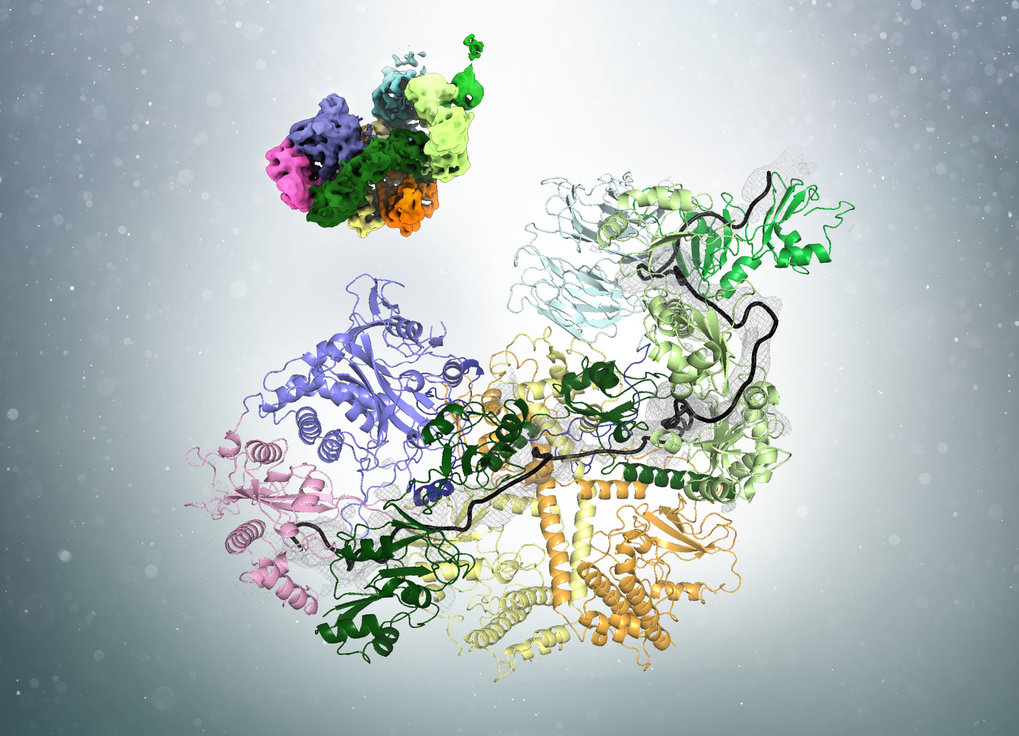
In a recent publication in the journal Cell, researchers led by Elena Conti at the Max Planck Institute of Biochemistry (MPIB) in Martinsried have demonstrated how poly(A)-tail shortening is controlled. “The poly(A)-tail is synthesized to a length much longer than we eventually find in cells. The presence of a poly(A)-tail-trimming mechanism has therefore been long suggested but the details were unclear”, says Ingmar Schäfer, a postdoctoral researcher in Elena Conti’s Department and first author of the study. The researchers now solved the structure of the involved components and show how measuring the length and trimming of the poly(A)-tail are coupled. The process requires the interplay of three components: the poly(A)-tail, poly(A)-tail-binding proteins (PABP) and the Pan2-Pan3 complex of deadenylases.
Using cryo-electron microscopy (cryo-EM), the researchers found that PABP forms arches that cover approximately 25 to 30 bases of the poly(A)-tail. Ingmar Schäfer uses an analogy from everyday life to illustrate the process: “Before cutting hair, a hairdresser uses his fingers to physically determine how much of the hair will be left standing.” Similarly, the PABP arches act as a rulers to determine the length of the poly(A)-tail. “But unlike our hair at the hairdresser, the poly(A)-tail is not trimmed with one cut but rather ‘nibbled off’ from the end by the deadenylase enzyme.”
Interestingly, the number of bound PABP alters the affinity of the deadenylase for the poly(A)-tail. In yeast, the most common poly(A)-tail length is around 30 nucleotides. On longer poly(A)-tails, several PABP can be bound and the adenine chain is quickly degraded. Shorter poly(A)-tails approaching the optimal length can only bind one ruler protein. This corresponds with a lower affinity for the deadenylases. “Hence, PABP is not only the tool measuring the poly(A)-tail, but also acts as a brake on the deadenylases when the optimal length is reached”, explains Schäfer. Eventually, when the mRNA is no longer needed, it is further degraded by a different set of deadenylases.
Polyadenylation is a basic principle of cell biology used to control mRNA stability and protein synthesis. “The structure of the poly(A)-tail ‘trimming tool’ will impact research on a broad variety of aspects of cell biology”, puts MPIB Director Elena Conti the study in a larger context. She highlights that the mechanism is highly conserved between yeasts and humans. “Now that we have solved the structure of the deadenylation machinery in yeast, we want to understand how the system works in human cells, where the optimal poly(A)-tail length differs from yeast.”
read more



